IMGT, the international ImMunoGeneTics information system®: a standardized approach for immunogenetics and immunoinformatics
- Research
- Open Access
IMGT, the international ImMunoGeneTics information system®: a standardized approach for immunogenetics and immunoinformatics
- Marie-Paule Lefranc1_3Email author
- Received: 21 June 2005
- Accepted: 20 September 2005
- Published: 20 September 2005
Abstract
IMGT, the international ImMunoGeneTics information system® http://imgt.cines.fr http://imgt.cines.fr, was created in 1989 by the Laboratoire d'ImmunoGénétique Moléculaire (LIGM)http://imgt.cines.fr/textes/IMGTinformation/LIGM.html (Université Montpellier II and CNRS) at Montpellier, France. IMGT is a high quality integrated knowledge resource specialized in immunoglobulins (IG), T cell receptors (TR), major histocompatibility complex (MHC) of human and other vertebrates, and related proteins of the immune system (RPI) of any species which belong to the immunoglobulin superfamily (IgSF) and to the MHC superfamily (MhcSF). IMGT consists of five databases, ten on-line tools and more than 8,000 HTML pages of Web resources. IMGT provides a common access to standardized data from genome, genetics, proteome and three-dimensional structures. The accuracy and the consistency of IMGT data are based on IMGT-ONTOLOGY, a semantic specification of terms to be used in immunogenetics and immunoinformatics. IMGT-ONTOLOGY comprises six main concepts: IDENTIFICATION, CLASSIFICATION, DESCRIPTION, NUMEROTATION, ORIENTATION and OBTENTION. Based on these concepts, the controlled vocabulary and the annotation rules necessary for the immunogenetics data identification, classification, description and numbering and for the management of IMGT knowledge are defined in the IMGT Scientific charthttp://imgt.cines.fr/textes/IMGTScientificChart/. IMGT is the international reference in immunogenetics and immunoinformatics for medical research (repertoire analysis of the IG antibody sites and of the TR recognition sites in autoimmune and infectious diseases, AIDS, leukemias, lymphomas, myelomas), veterinary research (IG and TR repertoires in farm and wild life species), genome diversity and genome evolution studies of the adaptive immune responses, biotechnology related to antibody engineering (single chain Fragment variable (scFv), phage displays, combinatorial libraries, chimeric, humanized and human antibodies), diagnostics (detection and follow up of residual diseases) and therapeutical approaches (grafts, immunotherapy, vaccinology). IMGT is freely available at http://imgt.cines.fr.
- IMGT
- antibody
- immunoglobulin
- T cell receptor
- superfamily
- MHC
- HLA
- ontology
- database
- information system
- knowledge resource
- immunoinformatics
- immunogenetics
- Collier de Perles
- three-dimensional
- 3D structure
- polymorphism
- annotation
Introduction
IMGT, the international ImMunoGeneTics information system® http://imgt.cines.fr http://imgt.cines.fr[1, 2], was created in 1989, by Marie-Paule Lefranchttp://imgt.cines.fr/textes/IMGTinformation/CVMPL.html, at the Laboratoire d'ImmunoGénétique Moléculaire (LIGM)http://imgt.cines.fr/textes/IMGTinformation/LIGM.html (Université Montpellier II and CNRS) at Montpellier, France, in order to standardize and manage the complexity of the immunogenetics data. Fifteen years later, IMGT is the international reference in immunogenetics and immunoinformatics, and provides a high quality integrated knowledge resource, specialized in the immunoglobulins (IG)http://imgt.cines.fr/textes/IMGTindex/antibodies.html and T cell receptors (TR)http://imgt.cines.fr/textes/IMGTindex/TR.html, major histocompatibility complex (MHC) of human and other vertebrates, and related proteins of the immune systems (RPI) of any species which belong to the immunoglobulin superfamily (IgSF)http://imgt.cines.fr/textes/IMGTindex/superfamily.html and to the MHC superfamily (MhcSF)http://imgt.cines.fr/textes/IMGTindex/superfamily.html[3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13]. The number of potential protein forms of the antigen receptors, IG and TRhttp://imgt.cines.fr/textes/IMGTindex/TR.html, is almost unlimited. The potential repertoire of each individual is estimated to comprise about 1012 different IG (or antibodies) and TR, and the limiting factor is only the number of B and T cells that an organism is genetically programmed to produce. This huge diversity is inherent to the particularly complex and unique molecular synthesishttp://imgt.cines.fr/textes/IMGTindex/synthese.html and genetics of the antigen receptor chains. This includes biological mechanisms such as DNA molecular rearrangements in multiple loci (three for IG and four for TR in humans) located on different chromosomes (four in humans), nucleotide deletions and insertions at the rearrangement junctions (or N-diversity), and somatic hypermutations in the IG loci (see FactsBookshttp://imgt.cines.fr/textes/IMGTindex/factsbook.html[3, 4] for review). Although IMGT was initially implemented for the IG, TR and MHC of human and other vertebrates [6], data and knowledge management standardization, based on the IMGT unique numbering [14, 15, 16, 17, 18, 19], has now been extended to the IgSF [15, 16, 17, 20, 21, 22] and MhcSF [18, 23, 24] of any species. Thus, standardization in IMGT contributed to data enhancement of the system and new expertised data concepts were readily incorporated.
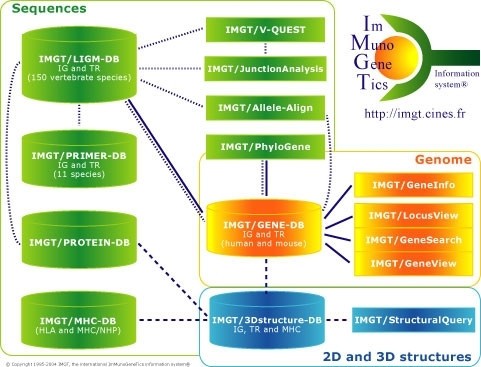
IMGT, the international ImMunoGeneTics information system® http://imgt.cines.fr. Databases and tools for sequences, genes and structures are in green, yellow and blue, respectively. The IMGT Repertoire and other Web resources are not shown. Interactions in the genetics, genomics and structural approaches are represented with dotted, continuous and broken lines, respectively.
IMGT-ONTOLOGY concepts and IMGT Scientific chart rules
IMGT-ONTOLOGY concepts, IMGT Scientific chart rules and examples of IMGT expertised data concepts.
The IMGT Scientific chart rules, based on the IMGT-ONTOLOGY concepts [5], are used in the three major IMGT biological approaches, genomics, genetics and structural approaches [2], and corresponding data (Genes, Sequences, 3D structures) are available in the IMGT components (databases, tools and Web resources) [1, 7, 8, 9, 10, 11, 12, 13].
IMGT sequence databases, tools and Web resources
The IMGT sequence databases, sequence analysis tools and Web resources
IMGT sequence databases
IMGT/LIGM-DB
IMGT/LIGM-DB is the comprehensive IMGT database of IG and TR nucleotide sequences from human and other vertebrate species, with translation for fully annotated sequences [7]. It was created in 1989 by LIGM (Montpellier, France), and is on the Web since July 1995 [6]. In August 2005, IMGT/LIGM-DB contained more than 96,500 sequences of 150 vertebrate species [7]. The unique source of data for IMGT/LIGM-DB is EMBL, which shares data with the other two generalist databases GenBank and DNA DataBank of Japan (DDBJ). Based on expert analysis, specific detailed annotations are added to IMGT flat files. The annotation procedure includes the IDENTIFICATION of the sequences, the CLASSIFICATION of the IG and TR genes and alleles, and the DESCRIPTION of all IG and TR specific and constitutive motifs within the nucleotide sequences. The Web interface allows searches according to immunogenetic specific criteria and is easy to use without any knowledge in a computing language. Selection is displayed at the top of the resulting sequences pages, so the users can check their own queries. Users have the possibility to modify their request or consult the results with a choice of nine possibilities. The IMGT/LIGM-DB annotations (gene and allele name assignment, labels) allow data retrieval not only from IMGT/LIGM-DB, but also from other IMGT databases. Thus, the IMGT/LIGM-DB accession numbers of the cDNA expressed sequences for each human and mouse IG and TR gene are available, with direct links to IMGT/LIGM-DB, in the IMGT/GENE-DB entries. IMGT/LIGM-DB data are also distributed by anonymous FTP servers at CINES ftp://ftp.cines.fr/IMGT/ and EBI ftp://ftp.ebi.ac.uk/pub/databases/imgt/ and from many Sequence Retrieval System (SRS) sites http://imgt.cines.fr/textes/IMGTotheraccesses.html. IMGT/LIGM-DB can be searched by BLAST or FASTA on different servers (EBI, IGH, INFOBIOGEN, Institut Pasteur, etc.).
IMGT/PRIMER-DB
IMGT/PRIMER-DB[1] is the IMGT oligonucleotide primer database for IG and TR, created by LIGM, Montpellier in collaboration with EUROGENTEC S.A., Belgium, on the Web since February 2002. In August 2005, IMGT/PRIMER-DB contained 1,827 entries. IMGT/PRIMER-DB provides standardized information on oligonucleotides (or Primers) and combinations of primers (Sets, Couples) for IG and TR. These primers are useful for combinatorial library constructions, scFv, phage display or microarray technologies. The IMGT Primer cards are linked to the IMGT/LIGM-DB flat files, IMGT Colliers de Perles and IMGT Alignments of alleles (IMGT Repertoire) of the IMGT/LIGM-DB reference sequence used for the primer description.
IMGT/MHC-DB
IMGT/MHC-DB[28] comprises databases hosted at EBI and includes a database of human MHC allele sequences or IMGT/MHC-HLA, developed by Cancer Research UK and maintained by ANRI, London, UK, on the Web since December 1998, and a database of MHC sequences from non human primates IMGT/MHC-NHP, curated by BPRC, The Netherlands, on the Web since April 2002.
IMGT sequence analysis tools
The IMGT sequence analysis tools comprise IMGT/V-QUEST[10], for the identification of the V, D and J genes and of their mutations, IMGT/JunctionAnalysis[11] for the analysis of the V-J and V-D-J junctions which confer the antigen receptor specificity, IMGT/Allele-Align for the detection of polymorphisms, and IMGT/PhyloGene[12] for gene evolution analyses.
IMGT/V-QUEST
IMGT/V-QUEST (V-QUEry and STandardization) is an integrated software for IG and TR [10]. This tool, easy to use, analyses an input IG or TR germline or rearranged variable nucleotide sequence. The IMGT/V-QUEST results comprise the identification of the V, D and J genes and alleles and the nucleotide alignments by comparison with sequences from the IMGT reference directoryhttp://imgt.cines.fr/textes/IMGTindex/directory.html, the FR-IMGT and CDR-IMGT delimitations based on the IMGT unique numbering, the translation of the input sequence, the display of nucleotide and amino acid mutations compared to the closest IMGT reference sequence, the identification of the JUNCTION and results from IMGT/JunctionAnalysis (default option), and the two-dimensional (2D) IMGT Collier de Perles representation of the V-REGION [10] ("IMGT/V-QUEST output" in IMGT/V-QUEST Documentationhttp://imgt.cines.fr/textes/vquest/imgtdnaplot.html).
IMGT/JunctionAnalysis
IMGT/JunctionAnalysishttp://imgt.cines.fr/cgi-bin/IMGTjcta.jv?livret=0[11] is a tool, complementary to IMGT/V-QUEST, which provides a thorough analysis of the V-J and V-D-J junction of IG and TR rearranged genes. IMGT/JunctionAnalysis identifies the D-GENEs and alleles involved in the IGH, TRB and TRD V-D-J rearrangements by comparison with the IMGT reference directory, and delimits precisely the P, N and D regions [11] ("IMGT/JunctionAnalysis output results" in IMGT/JunctionAnalysis Documentationhttp://imgt.cines.fr/v4/JCTA/Documentation.html). Several hundreds of junction sequences can be analysed simultaneously.
IMGT/Allele-Align
IMGT/Allele-Align is used for the detection of polymorphisms. It allows the comparison of two alleles highlighting the nucleotide and amino acid differences.
IMGT/PhyloGene
IMGT/PhyloGene[12] is an easy to use tool for phylogenetic analysis of variable region (V-REGION) and constant domain (C-DOMAIN) sequences. This tool is particularly useful in developmental and comparative immunology. The users can analyse their own sequences by comparing with the IMGT standardized reference sequences for human and mouse IG and TR [12] (IMGT/PhyloGene Documentationhttp://imgt3d.igh.cnrs.fr/phylogene/IMGTPhylogenyDoc.html).
IMGT sequence Web resources
The IMGT sequence Web resources are compiled in the IMGT Repertoire "Proteins and alleles" section that include Alignments of alleles, Proteins displays, Tables of alleles, Allotypes, Isotypes, etc. (Table 2). Standardized IMGT criteria for amino acid sequence analysis are described in [31].
IMGT gene databases, tools and Web resources
The IMGT gene database, genome analysis tools and Web resources
IMGT/GENE-DB, the IMGT gene database
Genomic data are managed in IMGT/GENE-DB, which is the comprehensive IMGT genome database [8]. IMGT/GENE-DB, created by LIGM (Montpellier, France) is on the Web since January 2003. In August 2005, IMGT/GENE-DB contained 1,377 genes and 2,207 alleles (673 IG and TR genes and 1,209 alleles from Homo sapiens, and 704 IG and TR genes and 998 alleles from Mus musculus, Mus cookii, Mus pahari, Mus spretus, Mus saxicola, Mus minutoïdes). All the human and mouse IG and TR genes are available in IMGT/GENE-DB. Based on the IMGT CLASSIFICATION concept, all the human IMGT gene names [3, 4] were approved by the Human Genome Organisation (HUGO) Nomenclature Committee HGNC in 1999 [27], and entered in IMGT/GENE-DB [8], Genome DataBase GDB (Canada) [32], LocusLink and Entrez Gene at NCBI (USA) [33], and GeneCards [34]. Reciprocal links exist between IMGT/GENE-DB, and the generalist nomenclature (HGNC Genew) and genome databases (GDB, LocusLink and Entrez at NCBI, and GeneCards). All the mouse IG and TR gene names with IMGT reference sequences were provided by IMGT to HGNC and to the Mouse Genome Database (MGD) [35] in July 2002. Queries in IMGT/GENE-DB can be performed according to IG and TR gene classification criteria and IMGT reference sequences have been defined for each allele of each gene based on one or, whenever possible, several of the following criteria: germline sequence, first sequence published, longest sequence, mapped sequence [2]. IMGT/GENE-DB interacts dynamically with IMGT/LIGM-DB [7] to download and display gene-related sequence data. As an example ans as mentioned earlier, the IMGT/GENE-DB entries provide the IMGT/LIGM-DB accession numbers of the IG and TR cDNA sequences which contain a given V, D, J or C gene. This is the first example of an interaction between IMGT databases using the CLASSIFICATION concept.
IMGT gene analysis tools
The IMGT gene analysis tools comprise IMGT/LocusView, IMGT/GeneView, IMGT/GeneSearch, IMGT/CloneSearch and IMGT/GeneInfo. IMGT/LocusView and IMGT/GeneView manage the locus organization and the gene location and provide the display of physical maps for the human IG, TR and MHC loci and for the mouse TRA/TRD locus. IMGT/LocusView allows to view genes in a locus and to zoom on a given area. IMGT/GeneView allows to view a given gene in a locus. IMGT/GeneSearch allows to search for genes in a locus based on IMGT gene names, functionality or localization on the chromosome. IMGT/CloneSearch provides information on the clones that were used to build the locus contigs displayed in IMGT/LocusView (accession numbers are from IMGT/LIGM-DB, gene names from IMGT/GENE-DB, and clone position and orientation, and overlapping clones from IMGT/LocusView). IMGT/GeneInfo[13] provides and displays information on the potential TR rearrangements in human and mouse.
IMGT gene Web resources
The IMGT gene Web resources are compiled in the IMGT Repertoire "Locus and genes" section that includes Chromosomal localizations, Locus representations, Locus description, Gene exon/intron organization, Gene exon/intron splicing sites, Gene tables, Potential germline repertoires, the complete lists of human and mouse IG and TR genes, and the correspondences between nomenclatures [3, 4] (Table 3). The IMGT Repertoire "Probes and RFLP" section provides additional data on gene insertion/deletion.
IMGT structure database, tool and Web resources
IMGT structure database, analysis tool and Web resources
|
IMGT structural database [1] |
IMGT structural analysis tool [1] |
IMGT Repertoirehttp://imgt.cines.fr/textes/IMGTindex/IMGTrep.html"2D and 3D structures"http://imgt.cines.fr/textes/IMGTrepertoire/2D-3Dstruct/ section [2] |
|---|---|---|
|
IMGT/3D structure-DBhttp://imgt.cines.fr/textes/IMGTindex/IMGT3Dstructure-db.html [15] |
IMGT/StructuralQuery [15] |
2D Colliers de Perles IG and TRhttp://imgt.cines.fr/textes/IMGTrepertoire/2D-3Dstruct/#1 [3, 4, 16, 17, 19] (1) 2D Colliers de Perles MHC [18, 36] 2D Colliers de Perles RPI [16-18, 21, 22, 24, 37] IMGT classes for amino acid characteristicshttp://imgt.cines.fr/textes/IMGTeducation/Aide-memoire/_UK/aminoacids/IMGTclasses.html [31] IMGT Colliers de Perles reference profiles [31] 3D representationshttp://imgt.cines.fr/textes/IMGTrepertoire/2D-3Dstruct/#3 (1) |
IMGT Colliers de Perles of a V-DOMAIN (A) and of a C-DOMAIN (B) (code PDB 1mcd in IMGT/3Dstructure-DB [9]). IMGT Colliers de Perles are shown on one layer (on the left hand side) and on two layers with hydrogen bonds (on the right hand side). (A) The IMGT Collier de Perles of a V-DOMAIN is based on the IMGT unique numbering for V-DOMAIN and V-LIKE-DOMAIN [16]. The CDR-IMGT are limited by amino acids shown in squares, which belong to the neighbouring FR-IMGT. The CDR3-IMGT extends from position 105 to position 117. CDR-IMGT regions are colored as follows on the IMGT site: CDR1-IMGT (blue), CDR2-IMGT (bright green), CDR3-IMGT (dark green) and hydrogen bonds are shown as green lines. (B) The IMGT Collier de Perles of a C-DOMAIN is based on the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN [17]. Amino acids are shown in the one-letter abbreviation. Arrows indicate the direction of the beta strands that form the two beta sheets of the immunoglobulin fold [3, 4]. Hatched circles correspond to missing positions according to the IMGT unique numbering [16, 17]. In the IMGT Collier de Perles on the IMGT Web site http://imgt.cines.fr hydrophobic amino acids (hydropathy index with positive value) and Tryptophan (W) found at a given position in more than 50 % of analysed IG and TR sequences are shown in blue, and all Proline (P) are shown in yellow.
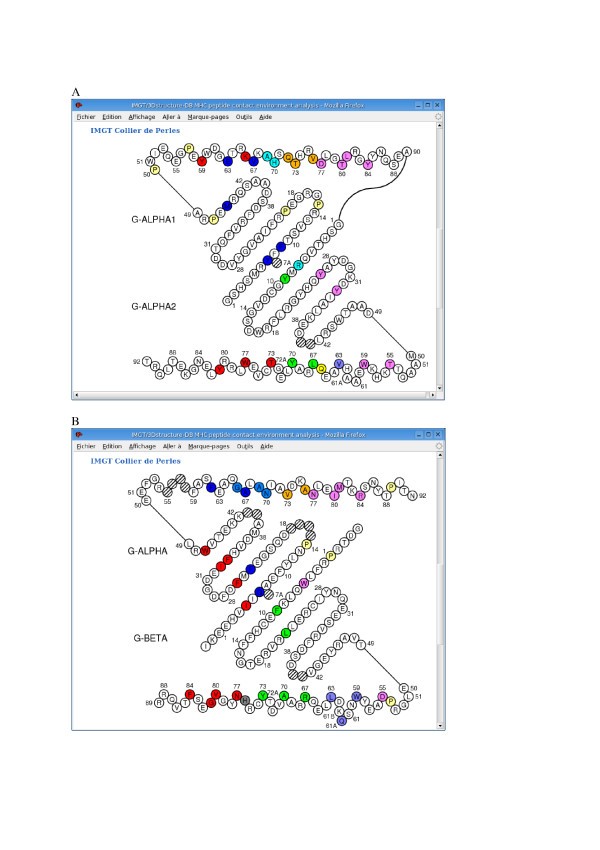
IMGT Colliers de Perles of the two G-DOMAINs of MHC class I (A) and of MHC class II (B) proteins (codes PDB 1bd2 and 1aqd, respectively, in IMGT/3Dstructure-DB [9]). The IMGT Collier de Perles of a G-DOMAIN is based on the IMGT unique numbering for G-DOMAIN and G-LIKE-DOMAIN [18]. (A) The two MHC-I G-DOMAINs, G-ALPHA1 (top) and G-ALPHA2 (bottom), form the groove of the MHC class I chain (I-ALPHA). (B) The two MHC-II G-DOMAINs, G-ALPHA (top) of the MHC class II alpha chain (II-ALPHA) and G-BETA (bottom) of the MHC class II beta chain (II-BETA), form the groove of the MHC class II protein [36]. Amino acids are shown in the one-letter abbreviation. Hatched circles correspond to missing positions according to the IMGT unique numbering [18]. Positions in colour correspond to the IMGT contact sites provided, for each peptide/MHC 3D structure, in IMGT/3Dstructure-DB [36].
IMGT/3Dstructure-DB, the IMGT 3D structure database
IMGT/3Dstructure-DB is the IMGT 3D structure database, created by LIGM, and on the Web since November 2001 [9]. In August 2005, IMGT/3Dstructure-DB contained 946 atomic coordinate files. IMGT/3Dstructure-DB comprises IG, TR, MHC and RPI with known 3D structures [9, 36, 37]. Coordinate files extracted from the Protein Data Bank (PDB) [38] are renumbered according to the standardized IMGT unique numbering [16, 17, 18]. The IMGT/3Dstructure-DB card provides, on-line, the complete information for each IMGT/3Dstructure-DB entry. The IMGT/3Dstructure-DB card shows a summary table and a menu that gives access to five sections: "Chain details", "Contact analysis", "Visualization with Jmol", "Renumbered file" and "References and links". The "Chain details" section provides chain description, IMGT gene and allele names, IMGT chain and domain labels, domain delimitations, amino acid positions according to the IMGT unique numbering, IMGT Colliers de Perles [16, 17, 18, 19]. The "Contact analysis" section provides contact types and categories between domains (in IMGT/3Dstructure-DB Domain contacts) and atom contacts at the residue and position level (in IMGT/3Dstructure-DB Residue@Position contacts) [37]. (IMGT/3Dstructure-DB Documentationhttp://imgt3d.igh.cnrs.fr/DOC/IMGT3DstructureDBHelp.shtml). The "Renumbered file" section downloadable provides renumbered IMGT/3Dstructure-DB flat files.
IMGT/StructuralQuery tool
The IMGT/StructuralQuery tool [9] analyses the interactions of the residues of the antigen receptors IG and TR, MHC, RPI, antigens and ligands. The contacts are described per domain (intra- and inter-domain contacts) and annotated in term of IMGT labels (chains, domain), positions (IMGT unique numbering), backbone or side-chain implication [37]. IMGT/StructuralQuery allows to retrieve the IMGT/3Dstructure-DB entries, based on specific structural characteristics: phi and psi angles, accessible surface area (ASA), amino acid type, distance in angstrom between amino acids, CDR-IMGT lengths.
IMGT structure Web resources
The IMGT stucture Web resources are compiled in the IMGT Repertoire "2D and 3D structures" section which includes 2D representations or IMGT Colliers de Perles [16, 17, 18, 19], 3D representations, FR-IMGT and CDR-IMGT lengths [16], amino acid chemical characteristics profiles [31], etc. In order to appropriately analyse the amino acid resemblances and differences between IG, TR, MHC and RPI chains, eleven IMGT classeshttp://imgt.cines.fr/textes/IMGTeducation/Aide-memoire/_UK/aminoacids/IMGTclasses.html were defined for the 'chemical characteristics' amino acid properties and used to set up IMGT Colliers de Perles reference profiles [31]. The IMGT Colliers de Perles reference profiles allow to easily compare amino acid properties at each position whatever the domain, the chain, the receptor or the species. The IG and TR variable and constant domains represent a privileged situation for the analysis of amino acid properties in relation with 3D structures, by the conservation of their 3D structure despite divergent amino acid sequences, and by the considerable amount of genomic (IMGT Repertoire), structural (IMGT/3Dstructure-DB) and functional data available. These data are not only useful to study mutations and allele polymorphisms, but are also needed to establish correlations between amino acids in the protein sequences and 3D structures and to determine amino acids potentially involved in the immunogenicity.
Conclusion
In order to allow any IMGT component to be automatically queried and to achieve a higher level of interoperability inside the IMGT information system and with other information systems, our current objectives include the modelling of the three major IMGT biological approaches, genomics, genetics and structural approaches, the analysis of the IMGT components (databases, tools and Web resources) in relation with the concepts, and the development of Web services http://www.w3.org/2002/ws/[2]. They are the first steps towards the implementation of IMGT-Choreography [2], which corresponds to the process of complex immunogenetics knowledge [25] and to the connection of treatments performed by the IMGT component Web services. IMGT-Choreography has for goal to combine and join the IMGT database queries and analysis tools. In order to keep only significant approaches, a rigorous analysis of the scientific standards [3, 4], of the biologist requests and of the clinician needs [39, 40, 41, 42] has been undertaken in the three main biological approaches: genomics, genetics and structural approaches. The design of IMGT-Choreography and the creation of dynamic interactions between the IMGT databases and tools, using the Web services and IMGT-ML, represent novel and major developments of IMGT, the international reference in immunogenetics and immunoinformatics. IMGT-Choreography enhances the dynamic interactions between the IMGT components to answer complex biological and clinical requests.
Since July 1995, IMGT has been available on the Web at http://imgt.cines.fr. IMGT has an exceptional response with more than 140,000 requests a month. The information is of much value to clinicians and biological scientists in general. IMGT databases, tools and Web resources are extensively queried and used by scientists from both academic and industrial laboratories, from very diverse research domains: (i) fundamental and medical research (repertoire analysis of the IG antibody sites and of the TR recognition sites in normal and pathological situations such as autoimmune diseases, infectious diseases, AIDS, leukemias, lymphomas, myelomas), (ii) veterinary research (IG and TR repertoires in farm and wild life species), (iii) genome diversity and genome evolution studies of the adaptive immune responses, (iv) structural evolution of the IgSF and MhcSF proteins, (v) biotechnology related to antibody engineering (single chain Fragment variable (scFv), phage displays, combinatorial libraries, chimeric, humanized and human antibodies), (vi) diagnostics (clonalities, detection and follow up of residual diseases) and (vii) therapeutical approaches (grafts, immunotherapy, vaccinology).
Citing IMGT
If you use IMGT databases, tools and/or Web resources, please cite [1] and this paper as references, and quote the IMGT Home page URL address, http://imgt.cines.fr.
Declarations
Acknowledgements
I am very grateful to Véronique Giudicelli, Chantal Ginestoux, Joumana Jabado-Michaloud, Géraldine Folch, Elodie Duprat, Denys Chaume, Quentin Kaas, and Gérard Lefranc for their expertise, constant motivation and helpful discussion. I am thankful to Wafae El Alaoui, Aurélie Frigoul, Lamia Zaghloul, François Ehrenmann, Arnaud Henry, Emmanuel-Jean Servier, our "2005" students, for their enthusiasm, and to the many IMGT users who have expressed their encouragement and support. IMGT is a registered mark of Centre National de la Recherche Scientifique (CNRS). IMGT has obtained the National Bioinformatics Platform RIO label since 2001 (CNRS, INSERM, CEA, INRA). IMGT was funded in part by the BIOMED1 (BIOCT930038), Biotechnology BIOTECH2 (BIO4CT960037) and 5th PCRDT Quality of Life and Management of Living Resources (QLG2-2000-01287) programmes of the European Union and received subventions from Association pour la Recherche sur le Cancer (ARC) and from the Génopole-Montpellier-Languedoc-Roussillon. IMGT is currently supported by the CNRS, the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche MENESR (Réseau National des Génopoles, Université Montpellier II Plan Pluri-Formation, Institut Universitaire de France, ACI-IMPBIO IMP82-2004 and BIOSTIC-LR2004 Région Languedoc-Roussillon) and GIS AGENAE (contrat AD2351 2005–2007).
Authors’ Affiliations
References
- Lefranc M-P, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clément O, Chaume D, Lefranc G: IMGT, the international ImMunoGeneTics information system ® . Nucl Acids Res 2005, 33:D593-D597. PMID: 15608269View ArticlePubMedGoogle Scholar
- Lefranc M-P, Clément O, Kaas Q, Duprat E, Chastellan P, Coelho I, Combres K, Ginestoux C, Giudicelli V, Chaume D, Lefranc G: IMGT-Choreography for immunogenetics and immunoinformatics. In Silico Biology 2004, 5:0006. Epub http://www.bioinfo.de/isb/2004/05/0006/, In Silico Biology, 2005, 5, 45–60. PMID: 15972004Google Scholar
- Lefranc M-P, Lefranc G: [http://imgt.cines.fr/textes/IMGTindex/factsbook.html] The Immunoglobulin FactsBook. Academic Press, London, UK; 2001, 458.Google Scholar
- Lefranc M-P, Lefranc G: [http://imgt.cines.fr/textes/IMGTindex/factsbook.html] The T cell receptor FactsBook. Academic Press, London, UK; 2001, 398.Google Scholar
- Giudicelli V, Lefranc M-P: Ontology for Immunogenetics: the IMGT-ONTOLOGY. Bioinformatics 1999, 12:1047–1054. PMID: 10745995View ArticleGoogle Scholar
- Lefranc M-P, Giudicelli V, Busin C, Malik A, Mougenot I, Déhais P, Chaume D: LIGM-DB/IMGT: an integrated database of Ig and TcR, part of the Immunogenetics database. Volume 764. Annals of the New York Academy of Sciences; 1995:47–49. PMID: 7486568Google Scholar
- Giudicelli V, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc M-P: IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res 2006.,34(January Database): Google Scholar
- Giudicelli V, Chaume D, Lefranc M-P: IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res 2005, 33:D256-D261. PMID: 15608191View ArticlePubMedGoogle Scholar
- Kaas Q, Ruiz M, Lefranc M-P: IMGT/3D structure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res 2004, 32:D208-D210. PMID: 14681396View ArticlePubMedGoogle Scholar
- Giudicelli V, Chaume D, Lefranc M-P: IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res 2004, 32:W435-W440. PMID: 15215425View ArticlePubMedGoogle Scholar
- Yousfi Monod M, Giudicelli V, Chaume D, Lefranc M-P: IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics 2004, 20:I379-I385. PMID: 15262823View ArticlePubMedGoogle Scholar
- Elemento O, Lefranc M-P: IMGT/PhyloGene: an on-line tool for comparative analysis of immunoglobulin and T cell receptor genes. Dev Comp Immunol 2003, 27:763–779. PMID: 12818634View ArticlePubMedGoogle Scholar
- Baum TP, Pasqual N, Thuderoz F, Hierle V, Chaume D, Lefranc M-P, Jouvin-Marche E, Marche PN, Demongeot J: IMGT/GeneInfo: enhancing V(D)J recombination database accessibility. Nucleic Acids Res 2004, 32:D51-D54. PMID: 14681357View ArticlePubMedGoogle Scholar
- Lefranc M-P: Unique database numbering system for immunogenetic analysis. Immunol Today 1997, 18:509. PMID: 9386342View ArticlePubMedGoogle Scholar
- Lefranc M-P: The IMGT unique numbering for Immunoglobulins, T cell receptors and Ig-like domains. The Immunologist 1999, 7:132–136.Google Scholar
- Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G: IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 2003, 27:55–77. PMID: 12477501View ArticlePubMedGoogle Scholar
- Lefranc M-P, Pommié C, Kaas Q, Duprat E, Bosc N, Guiraudou D, Jean C, Ruiz M, Da Piedade I, Rouard M, Foulquier E, Thouvenin V, Lefranc G: IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev Comp Immunol 2005, 29:185–203. PMID: 15572068View ArticlePubMedGoogle Scholar
- Lefranc M-P, Duprat E, Kaas Q, Tranne M, Thiriot A, Lefranc G: IMGT unique numbering for MHC groove G-DOMAIN and MHC superfamily (MhcSF) G-LIKE-DOMAIN. Dev Comp Immunol 2005, 29:917–938. PMID:15936075.View ArticlePubMedGoogle Scholar
- Ruiz M, Lefranc M-P: IMGT gene identification and Colliers de Perles of human immunoglobulins with known 3D structures. Immunogenetics 2002, 53:857–883. PMID: 11862387View ArticlePubMedGoogle Scholar
- Williams AF, Barclay AN: The immunoglobulin family: domains for cell surface recognition. Annu Rev Immunol 1988, 6:381–405. PMID: 3289571PubMedGoogle Scholar
- Duprat E, Kaas Q, Garelle V, Giudicelli V, Lefranc G, Lefranc M-P: IMGT standardization for alleles and mutations of the V-LIKE-DOMAINs and C-LIKE-DOMAINs of the immunoglobulin superfamily. Recent Res Devel Human Genet 2004, 2:111–136.Google Scholar
- Bertrand G, Duprat E, Lefranc M-P, Marti J, Coste J: Characterization of human FCGR3B*02 (HNA-1b, NA2) cDNAs and IMGT standardized description of FCGR3B alleles. Tissue Antigens 2004, 64:119–131. PMID: 15245367View ArticlePubMedGoogle Scholar
- Maenaka K, Jones EY: MHC superfamily structure and the immune system. Curr Opin Struct Biol 1999, 9:745–753. PMID: 10607669View ArticlePubMedGoogle Scholar
- Frigoul A, Lefranc M-P: MICA: standardized IMGT allele nomenclature, polymorphisms and diseases. Recent Res Devel Human Genet 2005, 3:95–145.Google Scholar
- Chaume D, Giudicelli V, Combres K, Ginestoux C, Lefranc M-P: IMGT-Choreography: processing of complex immunogenetics knowledge. Computational Methods in Systems Biology: International Conference CMSB Paris, France. In Lecture Notes in Computer Science. Edited by: Danos V, Schachter V. Springer-Verlag GmbH Berlin Heidelberg; 2004:73–84.Google Scholar
- Chaume D, Giudicelli V, Lefranc M-P: IMGT-ML a language for IMGT-ONTOLOGY and IMGT/LIGM-DB data. CORBA and XML: Towards a bioinformatics integrated network environment, Proceedings of NETTAB Network tools and applications in biology 2001, 71–75.Google Scholar
- Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S: Guidelines for human gene nomenclature. Genomics 2002, 79:464–470. PMID: 11944974View ArticlePubMedGoogle Scholar
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG: IMGT/HLA and IMGT/MHC sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 2003, 31:311–314. PMID: 12520010View ArticlePubMedGoogle Scholar
- Giudicelli V, Chaume D, Jabado-Michaloud J, Lefranc M-P: Immunogenetics sequence annotation: the strategy of IMGT based on IMGT-ONTOLOGY. In The XIXth International Congress of the European Federation for Medical Informatics, Geneva, Switzerland, Connecting Medical Informatics and Bio-informatics. Proceedings of the Medical Informatics Europe MIE. Volume 116. Edited by: Engelbrecht R, Geissbuhler A, Lovis C, Mihalas G. IOS Press, Technology and Informatics; 2005:3–8.Google Scholar
- Giudicelli V, Lefranc M-P: Interactive IMGT on-line tools for the analysis of immunoglobulin and T cell receptor repertoires. In New Research on Immunology. Edited by: Veskler BA. Nova Science; 2005:77–105.Google Scholar
- Pommié C, Sabatier S, Lefranc G, Lefranc M-P: IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J Mol Recognit 2004, 17:17–32. PMID: 14872534View ArticlePubMedGoogle Scholar
- Letovsky SI, Cottingham RW, Porter CJ, Li PW: GDB: the Human Genome Database. Nucleic Acids Res 1998, 26:94–99. PMID: 9399808View ArticlePubMedGoogle Scholar
- Pruitt KD, Maglott DR: RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res 2001, 29:137–140. PMID: 11125071View ArticlePubMedGoogle Scholar
- Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, Rosen N, Shmoish M, Peter Y, Glusman G, Feldmesser E, Adato A, Peter I, Khen M, Atarot T, Groner Y, Lancet D: Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res 2003, 31:142–146. PMID: 12519968View ArticlePubMedGoogle Scholar
- Blake JA, Richardson JE, Bult CJ, Kadin JA, Eppig JT, Mouse Genome Database Group: MGD: the Mouse Genome Database. Nucleic Acids Res 2003, 31:193–195. PMID: 12519980View ArticlePubMedGoogle Scholar
- Kaas Q, Duprat E, Le Tourneur G, Lefranc M-P: IMGT standardization for molecular characterization of the T cell receptor/peptide/MHC complexes. Springer, in press.Google Scholar
- Kaas Q, Lefranc M-P: Interactive IMGT on-line database and tool for the structural analysis of immunoglobulins, T cell receptors, MHC and related proteins of the immune system. Focus on Immunology Research, Nova Science, in press.Google Scholar
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res 2000, 28:235–242. PMID: 10592235View ArticlePubMedGoogle Scholar
- Chassagne S, Laffly E, Drouet E, Herodin F, Lefranc M-P, Thullier P: A high affinity macaque antibody Fab with human-like framework regions obtained from a small phage display immune library. Mol Immunol 2004, 41:539–546. PMID: 15183932View ArticlePubMedGoogle Scholar
- Laffly E, Danjou L, Condemine F, Vidal D, Drouet E, Lefranc M-P, Bottex C, Thullier P: Selection of a macaque Fab with human-like framework regions, high affinity, and that neutralizes the protective antigen (PA) of Bacillus anthracis . Antimicrob Agents Chemother 2005, 49:3414–3420. PMID: 16048955View ArticlePubMedGoogle Scholar
- Stamatopoulos K, Belessi C, Papadaki T, Kalagiakou E, Stavroyianni N, Douka V, Afendaki S, Saloum R, Parasi A, Anagnostou D, Laoutaris N, Fassas A, Anagnostopoulos A: Immunoglobulin heavy- and light-chain repertoire in Splenic Marginal Zone Lymphoma. Mol Med 2005, in press. PMID: 15706403Google Scholar
- Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stella S, Guida G, Michel A, Crespo M, Laoutaris N, Montserrat E, Anagnostopoulos A, Dighiero G, Fassas A, Caligaris-Cappio F, Davi F: Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3–21 gene. Blood 2005, 105:1678–1685. PMID: 15466924View ArticlePubMedGoogle Scholar
Copyright
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.